Quantitative Live-Cell Imaging Analysis
Interview with Ed Luther, Northeastern University in Boston
Since 2014, HoloMonitor and quantitative live-cell imaging analysis play an important role in advancing the scientific interests of researchers at Northeastern University.
The HoloMonitor live-cell imaging and analysis system provides a multitude of cellular parameters — reflecting cell health, proliferation, migration, differentiation and cell cycle phase, on a population or single-cell level. From just a single live-cell experiment, scientists at the Department of Pharmaceutical Sciences non-invasively study how treatments affect cellular response and behavior in cell-based models.
We reached out to the lab supervisor at the department, Ed Luther, to ask about their work. Ed has a long and passionate interest in live-cell imaging and cytometry. Over the years he has had the privilege to run core cytometry facilities at Harvard University and elsewhere. Today Luther is based in Boston Mass. at the Department of Pharmaceutical Sciences, Northeastern University.
Ed, how did you get involved in quantitative imaging and live-cell analysis?
Shortly after I started my work at Northeastern University (NEU), a collaborative agreement was signed with Phase Holographic Imaging (PHI), which brought us the new HoloMonitor. The introductory presentations piqued the interest of many researchers at the department.
What caught their interest back then?
We had done extensive work with the glioblastoma cells over the years, but it was always difficult because of their perceived high level of motility, which made standard assays such as wound healing ineffective.

Getting to work with HoloMonitor is a natural extension of my interests and expertise
What did this initial interest lead to?
A combination of features, i.e. label-free and quantitative live-cell analysis, ease of use and the ability to operate the instrument within a standard incubator environment, have rapidly established the HoloMonitor as a primary analytical workhorse. With HoloMonitor, meaningful results have been and are obtained in a short period of time.
Through the department’s activities, we now provide cytometry outreach programs, consultations on performing HoloMonitor experiments and analyzing the data. We promote the practice of label-free cytometry and strive to maintain a professional network serving as a center of knowledge and expertise. As evident from our publications and scientific presentations schedule, HoloMonitor results play an important role in advancing the scientific interests of NEU researchers.
How did you personally get involved with image-based cell analysis in the first place?
My academic background is actually in paleontology at the University of California, Berkeley. I was studying 2 billion years old cherts of the Gunflint Formation contained the oldest known fossil organisms at the time. The microscopy skills that I later developed led to my joining Professor Donald A. Glaser [Nobel Prize winner for the development of the bubble chamber], who at this time started developing his facility for Automated Experiments in Cell Biology. The prototype instrument had its own building with the instrument chamber, a very large room with laminar airflow and temperature control designed to operate at 37 degrees.
The label-free aspects of HoloMonitor and quantitative phase imaging really provide new capabilities that we did not have before
Tell us about your research interests?
My subsequent research area evolved to quantitative analysis of living cells and tissues. I have been doing this for many decades, both in academic institutions, operating core facilities, and in private industry. Platforms include live-cell microscopy, large scale automated microscopy, flow cytometry, large scale automated flow cytometry, laser scanning cytometry, and large-scale laser scanning cytometry for both cellular samples and tissues.
I am currently based at the Department of Pharmaceutical Sciences, Northeastern University in Boston. My role here is to establish the imaging cytometry core facilities for use of researchers, faculty and students. The department is located within the Bouve College of Health Sciences. While its mission is mostly educational it does contain and has access to many specialized research centers, such as the Center for Pharmaceutical Biotechnology and Nanomedicine. The center has a strong emphasis on nanotechnology, especially the development of complex drug delivery formulations that target the eradication of specific cancer cells but do no or minimal damage to other cell types within the host.
Can you tell us about your publication A Special Issue in Quantitative Image Analysis Applications of Label-Free, Quantitative Phase Holographic Imaging Cytometry to the Development of Multi-Specific Nanoscale Pharmaceutical Formulations?
We were quite well along in our application development, as well as publishing results in high impact journals when the call for papers went out for this special edition, so we wanted to chronicle our experience with quantitative phase imaging. We started with a brief overview of the HoloMonitor Mach-Zehnder optical system, including a general description of the image processing and analysis functions of the system, and then went on to describe our first major contribution to the field, the 4-dimensional imaging. Each of these pictures is worth more than the proverbial thousand words in the value of their information conveyance.
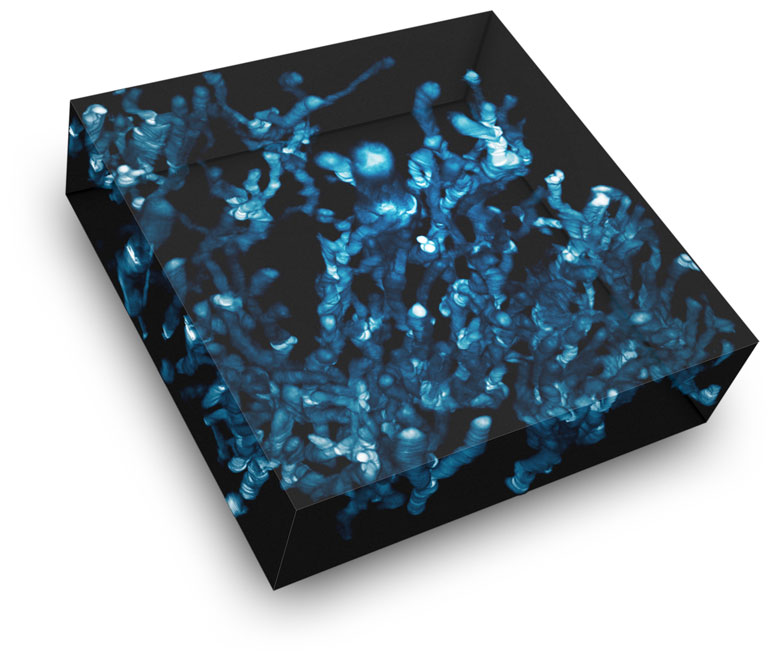
The image, conceptualized by Ed Luther, shows a proliferating and migrating cell population. The spectacular live-cell image was created by overlaying HoloMonitor time-lapse images in a single image, where time is shown along the z-axis.
A set of images was taken, with the goal of finding the largest cell possible for long-term tracking. The cell grew to giant proportions as was described in the paper, and although the process was barely heard of at the time, we had stumbled onto Neosis – a survival method where giant polyploid cells autophagize and reprocess DNA to form tumor stem cells. We then verified that label-free imaging is capable of providing the same results that we would obtain when cell lines are treated with a panel of standard toxins.
The rest of the paper consists of synopses of publications in refereed journals emphasizing the portions of the studies that were done using the HoloMonitor. In the macrophage polarization study produced with members of Professor Amiji’s laboratory, we showed how macrophage polarization can be controlled by chemical stimuli. Another example describing a combinatorial liposomal formulation to overcome multidrug resistance in ovarian cancer cells used 4D live-cell imaging to demonstrate the effects of the formulation components over time.
Luther et al., Applications of label‐free, quantitative phase holographic imaging cytometry to the development of multi‐specific nanoscale pharmaceutical formulations, Cytometry Part A (2017)
Luther et al., Comparison of the effect of pharmaceutical compounds on tumor cells in 2D and 3D in vitro models using label-free, quantitative 4 dimensional holographic imaging, AACR-NCI-EORTC (2015)
Can you tell us about your latest publication Advancing methods for the analysis of glioblastoma cell motion using quantitative time-lapse holographic imaging and cellular tomography?
We were invited to present our work at the 2018 SPIE Photonics West meeting in San Francisco. As I mentioned earlier, we had previously done extensive work with the glioblastoma cells, but it was always difficult because of their perceived high level of motility, making the standard wound healing assay ineffective. The first significant part of the work involved characterizing and developing new HoloMonitor algorithms for documenting their movement patterns.
The second step necessitated us to have access to a cellular tomography platform with nano-level optical resolution, which we included in our investigations. We were able to visualize the complicated movements and morphology changes of the cells, but also the movements of the organelles within the cells such as the mitochondria, their fission and tunneling activities. This higher level of resolution leads to new insights in intercellular communication and social cellular networks. It had previously been shown that glioblastoma cells contain a subset of tumor stem cells, and we believe we have developed a method for virtually isolating them.
Once we learned what to look for, we were able to devise methodologies using HoloMonitor to locate the cells of interest at lower resolutions. Why did we need to adopt the application to HoloMonitor? The answer to this question is the ability to evaluate much larger numbers of living cells and obtain quantitative data. We can now run kinetic assays of the cell movement, such as wound healing assays, cellular expansion assays, as well as chemotactic assays to evaluate the effects of our formulations.
Luther et al., Advancing methods for the analysis of glioblastoma cell motion using quantitative time-lapse holographic imaging and cellular tomography, Proc. SPIE (2019)
HoloMonitor live-cell assays are cell-based, and the far more information can be extracted from individual cells
How has HoloMonitor helped you with your research?
My research concentrates on developing applications for new instrument platforms. My primary objectives are validation of analytical methods and developing robust and accurate live-cell analysis routines to assure that our researchers produce quality data.
Quantitative time-imaging imaging of living cells brings several new possibilities. One of the most important features of HoloMonitor is the ability to analyze entire cell populations and the individual cells within them. The time-lapse aspect is the major driver of this, but it must be pointed out that video microscopy has been along for a long time, with the work of Shinya Inoue’ of Woods Hole Biological Laboratory being an excellent example.
However, most of the prior label-free live-cell imaging methodologies, such as brightfield and standard phase contrast microscopy, images suffer from poor contrast, making segmentation and analysis difficult. HoloMonitor images are darkfield, automatically focused, and are perfectly amenable to post-processing techniques. I primarily use readily available programs such as Image J, Microsoft Excel, and Windows Media Player.
Our HoloMonitor glioblastoma cell studies confirm the emerging trend that tumors are not merely collections of cells with defined boundaries, but are more like organ systems, with continuous and robust movements of subcellular moieties between the individual members.
Our live-cell studies of giant polyploid cells show processes like neosis where autophagy and reprocessing of genetic material to form potential tumor stem cells. In the process, many long-dormant ancient genes can be reactivated, and in effect, reverse or de-evolution.
Would you have been able to get the same quantitative results without HoloMonitor?
Typical assays in pharmaceutical sciences are done in bulk with the results presented as averages of the whole population. The HoloMonitor live-cell imaging assays are time-lapse-based, and far more information can be extracted from individual cells. In our first HoloMonitor breakthrough, we were witnessing giant cells and monitored their life cycles over many generations.
Polyploidy is emerging as a possible cause and not just an effect of cancer cell evolution. Processes such as autophagy and cell component recycling and resurrection of ancient genes are becoming vital to the understanding of cancer cell etiology. Combined with the increasing awareness of the effects of the tumor microenvironment, we are able to devise more effective anti-cancer agents.
I also recall Peter Egelberg’s [CEO, Phase Holographic Imaging] conversation during his visit with the Distinguished Professor Vladimir Torchilin who responded to a similar question from Peter: “While we have multiple alternative methodologies to obtain the information we need, with HoloMonitor we get better presentations of the data and are able to publish in better journals, and become competitive in obtaining grants”.
HoloMonitor brings a number of new possibilities. One of the most important features is the ability to analyze entire cell populations and the individual cells within them
The future with HoloMonitor – what potential do you see in the near future?
I manage a shared research facility with many different cytometry and microscopy instruments. I have been following the development of the HoloMonitor from the early stages of its commercial release. The label-free aspects of quantitative phase imaging really provide new capabilities that we did not have before. I have put in enough time working in the field to declare with some degree of credibility in professing that one or more HoloMonitor units have a place in any high-level cellular imaging facility.
My forward-thinking is that quantitative live-cell phase imaging has many attributes that could potentially be useful in the clinical diagnostic realm. This will most likely require many years of validation, but I envision that PHI at some point will be involved in clinical applications. I can think of a potential diagnostic application where label-free imaging may be employed – the field of Interventional Radiology. Needle aspirates and core biopsies have a long history in triage and diagnosis, mostly in part of their minimal invasiveness and lack of collateral damage to the patient.
With HoloMonitor technology no labeling is required. Preliminary readouts can be obtained instantaneously, resulting in a convenient evaluation of aspirates or tissues. Getting results as soon as possible is of interest, especially when the patient is tying up a very expensive piece of equipment. In standard radiology suites, patients are usually cycled through in a constant stream to maximize the return from the investments.
What are your most significant developments in quantitative live-cell imaging and cellular analysis?
Flow cytometry is generally regarded as the first quantitative cell-based technology, and its successor, laser scanning cytometry, is the first quantitative imaging technology. In advancing our analytical method we decided to take advantage of easy access to the time-lapse image sequences and came up with an idea to analyze the cell thickness parameter over time as an image stack.
We can now comprehensively visualize cell cycle information, expansion of cell clones, cell death process, cell motility, cellular adhesion, cellular degradation, and debris within the sample area – every real and quantifiable event, as well as the unquantifiable elements of the environment.
When commercial flow cytometers were first introduced, Ted Young proposed the Kolmogorov – Smirnov (KS) statistical test to determine the statistical significance between two flow cytometry histograms. Ken’s laboratory was where I was introduced to clinical flow cytometry. I have continued to use and develop new versions of the KS test to this day, now applying it HoloMonitor data.