Holographic Microscopy
Holographic microscopy is the most common form of quantitative phase imaging. The HoloMonitor® live cell imaging microscope employs digital holographic microscopy to allow non-invasive visualization and quantification of living cells without compromising cell integrity.
Conventional Holography
A traditional hologram is recorded on a photographic plate. The holographic image is created by illuminating the developed hologram with the laser that was used to create the hologram. This recreates the same light wavefield that originally came from the imaged object. The object appears to be 3-dimensional as each eye image a slightly different part of the wavefield, just like they would have done if the object was still there.
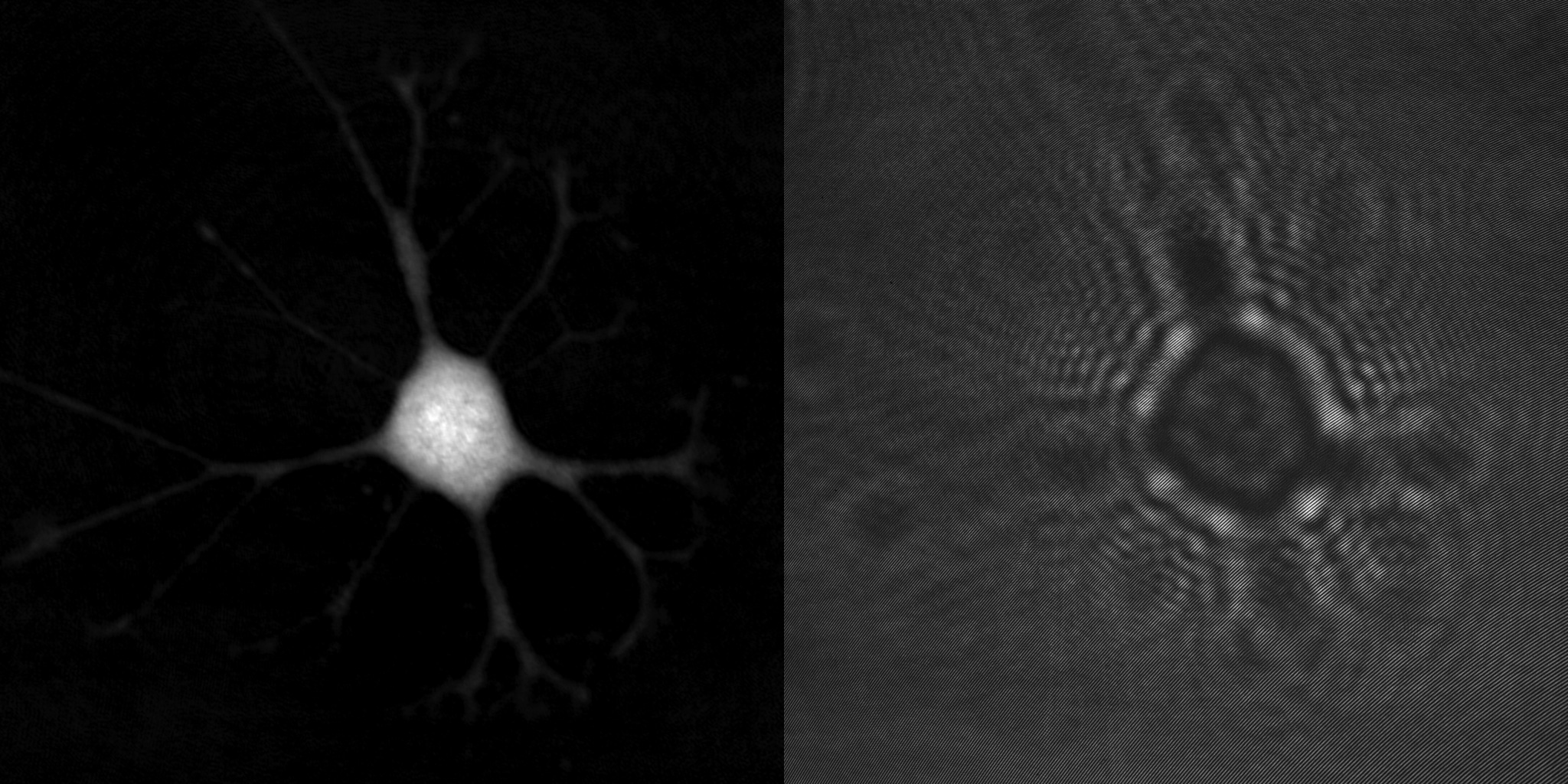
A cell phase image (left) and the hologram it was created from (right).
Digital Holography
The advent of high-resolution image sensors has made it possible to instead record a hologram digitally, allowing the holographic image to be created by a computer, rather than re-illuminating the developed hologram. The computer actually creates two images: an amplitude (intensity) and a phase image. As unstained cells are transparent, most of the information resides in the phase image.

When light waves interact they create an interference pattern, just like water waves do.
The Holographic Microscopy Principle
Holographic microscopy creates (quantitative) phase images by letting a sample beam and a reference beam interfere to create an interference pattern or hologram, as shown in the principle image below. The hologram is recorded by an image sensor and computer-processed to produce the final phase image.
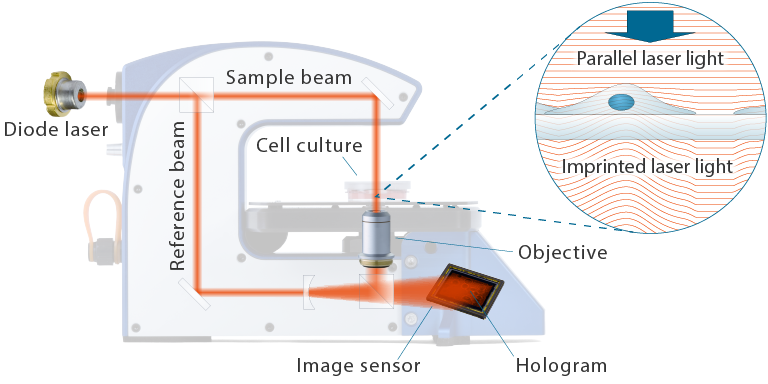
The principle design of HoloMonitor
It is useful to think of a phase image as a picture of the optical imprint created by the cells. When the illuminating sample beam passes through the sample, the sections of the beam that passes through the more optically dense cells are delayed in relation to the background. This shifts the phase of the parallel sample beam and imprints the morphology and 3-dimensional optical properties of the cells on the sample beam, similar to how beach waves are delayed and phase-shifted when they reach shallow water.

Shallow water imprinted beach waves
Holographic Microscopy References

Label-free High Temporal Resolution Assessment of Cell Proliferation Using Digital Holographic Microscopy
Authors: Birgit Janicke et al.
Journal: Cytometry Part A (2017)
Research Areas: Method development
Cell Lines: L929
Keywords: HoloMonitor M4, Cell proliferation, Digital holographic microscopy, Cell count, Noninvasive, Label-free, Image cytometry, Quantitative phase imaging, Time-lapse, Digital holography technology

Digital Holographic Microscopy for Non-invasive Monitoring of Cell Cycle Arrest in L929 Cells
Authors: Maria Falck Miniotis et al.
Journal: PLOS ONE (2014)
Research Areas: Method development
Cell Lines: L929
Keywords: Holomonitor M3, Cell morphology, Cell cycle and cell division, Flow cytometry, Cancer treatment, G1 phase, Colcemid treatment, Holographic microscopy, therapeutic drug
Cells and Holograms — Holograms and Digital Holographic Microscopy as a Tool to Study the Morphology of Living Cells
Authors: Kersti Alm et al.
Journal: Holography — Basic Principles and Contemporary Applications (2013)
Research Areas: Method development
Keywords: Digital holographic technology

Refractometry of Microscopic Objects With Digital Holography
Authors: Mats Gustafsson et al.
Journal: Applied Optics (2004)
Research Areas: Method development
Keywords: digital holographic imaging, effective refractive index, light intensity, phase contrast, phase imaging, refractive index, digital holography technology