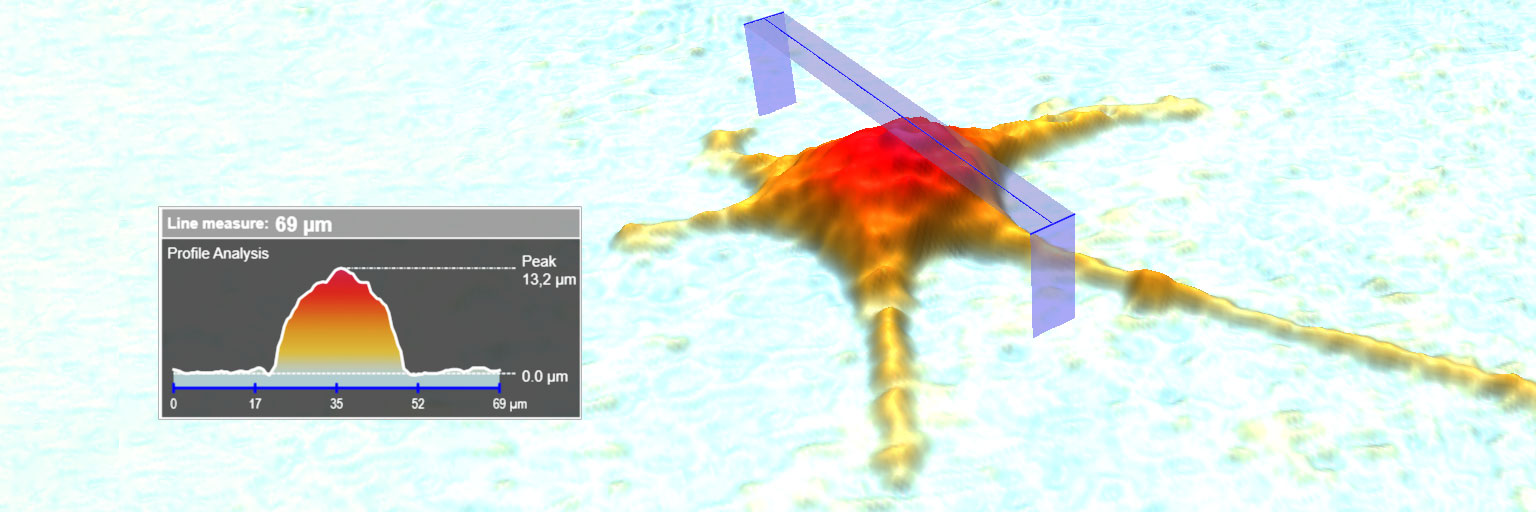
created using HoloMonitor®

Wound Healing With Single-cell Tracking
The HoloMonitor® Wound Healing Scratch Assay analyze in vitro cell migration. Cell migration speed is automatically determined in a straightforward way, directly in your incubator.
The in vitro wound healing assay, also known as scratch assay, is widely used to model in vivo cell migration. The method is based on quantifying the rate at which cells repopulate an artificial gap or scratch created in a confluent monolayer of cells.
Single-cell Tracking
The HoloMonitor incubator microscope provides an automated label-free wound healing assay, measuring scratch closure rate and cell front velocity. Additionally, cell tracking may be applied on selected cell front cells for detailed individual cell movement and morphology analysis.
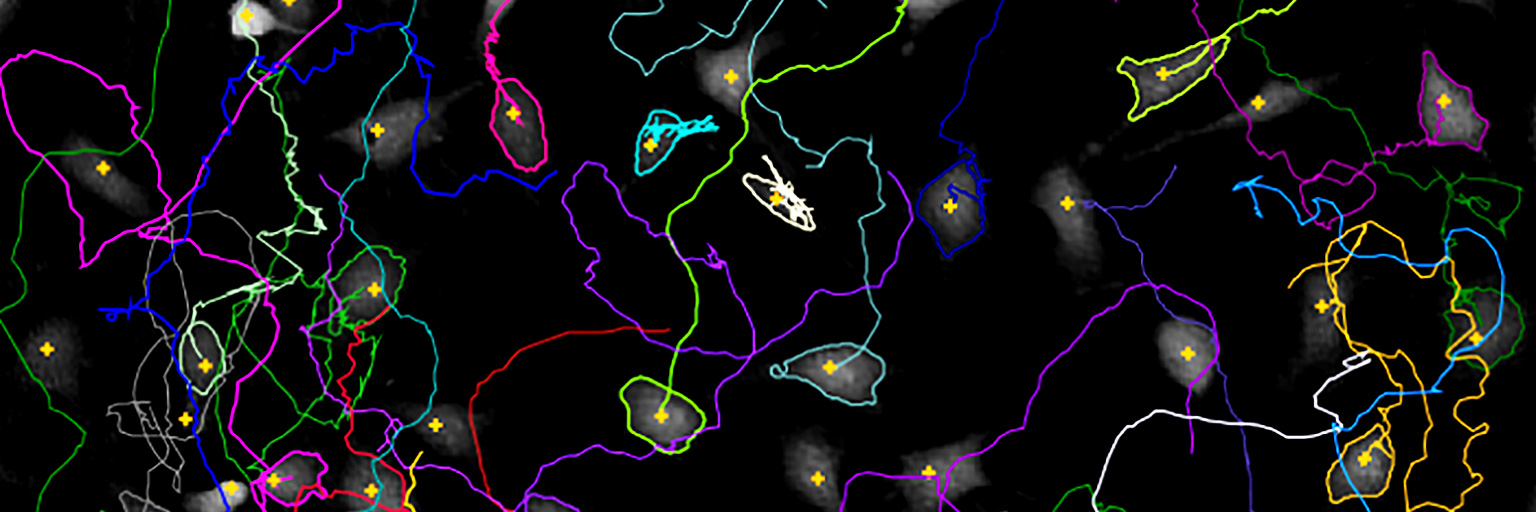
After recording the wound healing time-lapse image sequence, individual cells at the cell front may be tracked to determine their migration direction with the HoloMonitor Single-cell Tracking Assay. Detailed, non-biased data on migration into the wound area is then easily achieved. When preferred, further analyses can be undertaken for details on cell morphology changes, cell proliferation and cell division using the HoloMonitor Live Cell Assays.
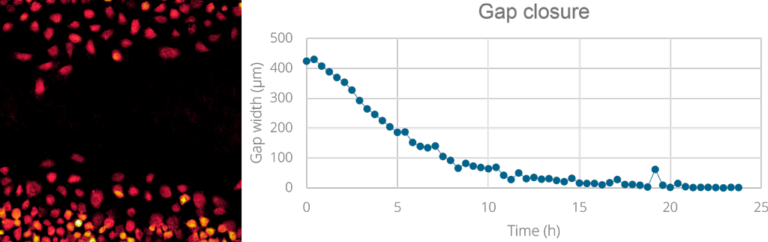
The HoloMonitor Wound Healing Assay provides kinetic gap closure analysis showing how gap width and cell covered area change over time.
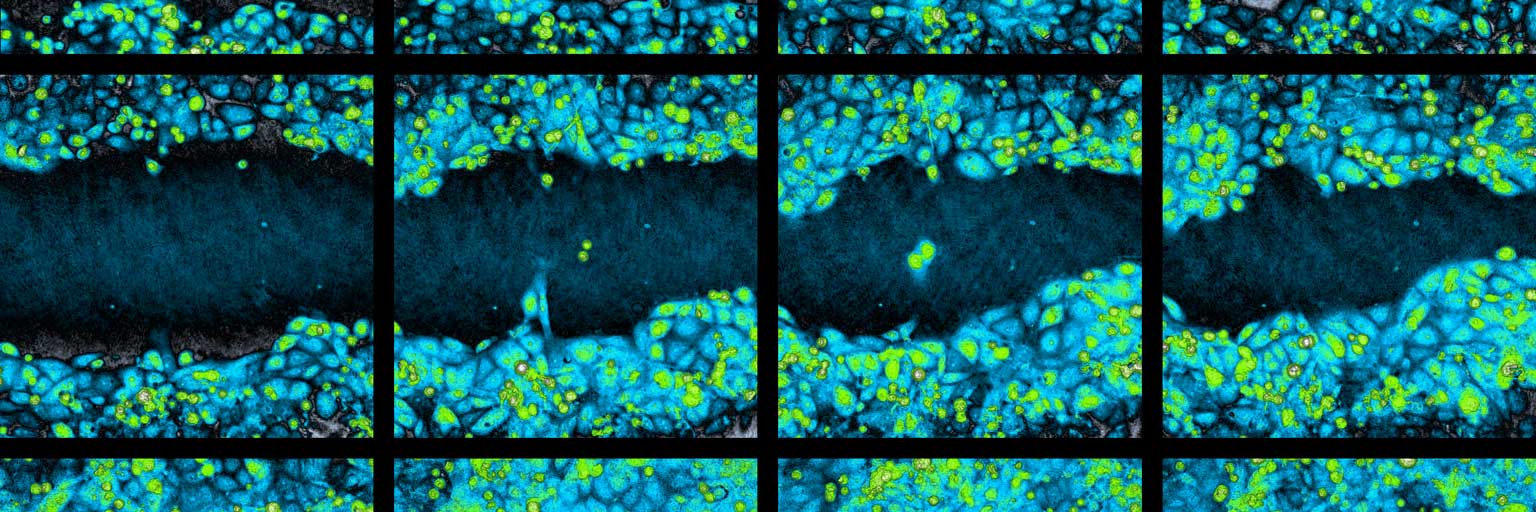
Videos showing gap closure over time as the cells fill the gap. Untreated cells are used and the gap is created using the ibidi® Culture-Insert 2 Well.
Tracked cell front cells, overlayed tracks (left), migration plot (right).
HoloMonitor® Wound Healing Assay — Get the Whole Picture
The assay is based on a reproducible method to create the gap, coupled with graphics and automated data analysis to provide gap closure rate. As with all HoloMonitor applications, it is cell-friendly and based on label-free live cell imaging.
- Cell front velocity
- Time-lapse videos showing how cells migrate into the gap
- Automatic and kinetic calculation of gap closure over time
- Single-cell tracking
Key References

Moving into a New Dimension: Tracking Migrating Cells with Digital Holographic Cytometry in 3D
Authors: Anette Görloff Wingren
Journal: Cytometry Part A (2018)
Research Areas: Cancer research
Keywords: HoloMonitor M4, cell motility, cell migration, cell morphology, cell tracking, cytometry, digital holography, migration, motility, quantitative phase imaging, scratch assay

Evaluation of Holographic Imaging Cytometer HoloMonitor M4 Motility Applications
Authors: Yuntian Zhang et al.
Journal: Cytometry Part A (2018)
Research Areas: Cancer research
Cell Lines: 1205Lu and WM793
Keywords: HoloMonitor M4, Cell motility, Cell migration, Cell morphology