10 Questions to Besa
Find the full interview below. Enjoy reading!
The HoloMonitor system supports our research by enabling detailed, non-invasive, and real-time monitoring of cell behaviors, which has been crucial for advancing our understanding of the cellular effects of potential anticancer compounds and the fundamental differences between normal and cancerous breast cells.
Q1: Besa, can you tell us more about yourself and your research focus?
A: I am an Assistant Professor of Biochemistry, deeply engaged in cancer research. My work primarily explores the molecular mechanisms of tumor development and progression, with a special focus on identifying novel therapeutic targets. Recently, I have been investigating the anti-tumorigenic effects of natural compounds on estrogen receptor-positive breast cancer cells. I am aiming to understand how these agents can modulate signaling pathways to inhibit cancer growth and metastasis. My research also extends to the impacts of environmental toxins on cell differentiation and development, providing insights into their roles in cancer and other diseases.
Q2: How did HoloMonitor help you address your research question or goal?
A: The HoloMonitor system has been instrumental in addressing our research goals by providing advanced capabilities for live-cell imaging and quantitative analysis. Specifically, its ability to perform holographic time-lapse microscopy allowed us to non-invasively monitor and analyze the dynamic cellular behaviors and morphological changes of breast cancer cells over extended periods. This technique was crucial in our study to assess the effects of the natural compounds in breast cancer cells.
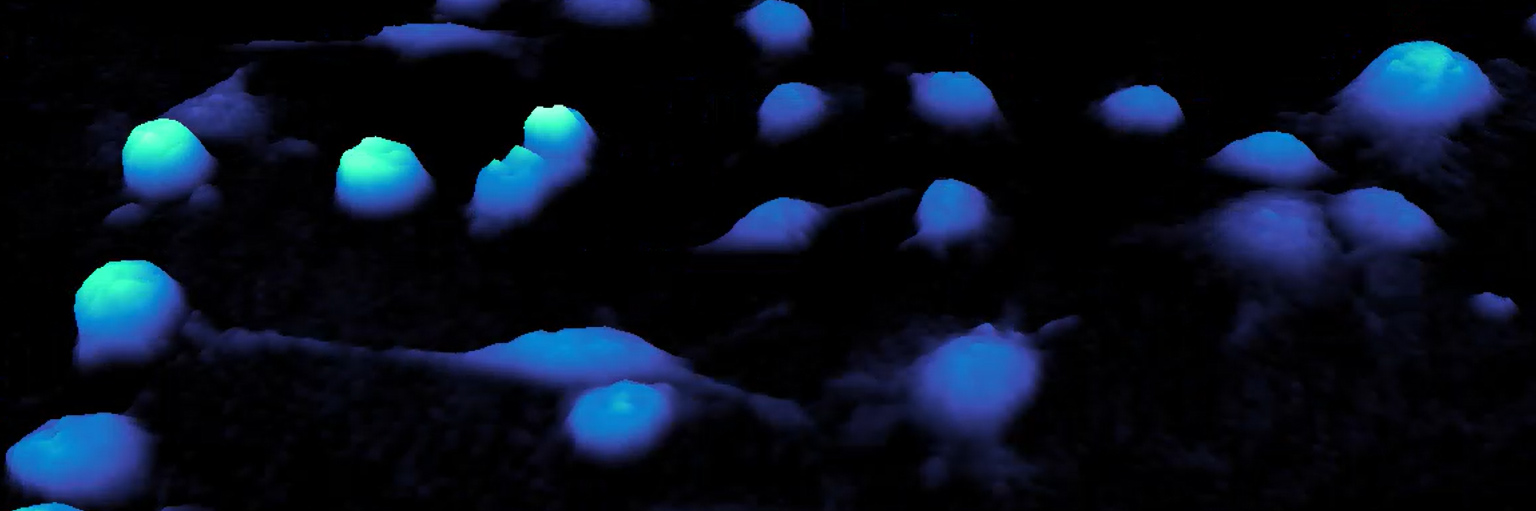
Using the HoloMonitor, we were able to capture detailed images every 10 minutes for 24 hours. We received a continuous, real-time view of cellular processes such as proliferation, motility, and morphological changes, and much more in response to treatment. The system’s software facilitated quantitative analysis of cell proliferation rates and detailed morphological parameters such as cell volume, area, and diameter. This allows us to generate comprehensive data sets that were statistically analyzed to identify significant changes in cell behavior.
Besa shares some of her MCF7 cell data
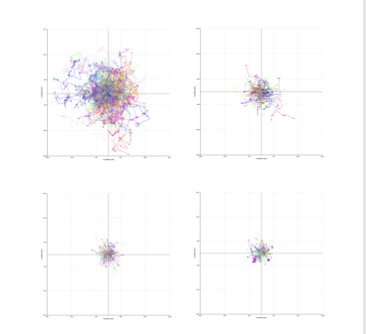
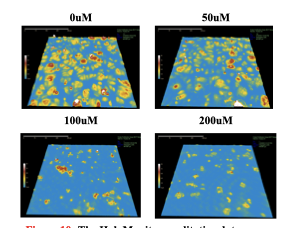
Furthermore, the label-free imaging capability of HoloMonitor ensured that our observations were not confounded by potential artifacts from dyes or labels, thus keeping cellular integrity and providing more physiologically relevant data. This feature was particularly valuable in our comparative analysis of the morphological features of normal breast and breast cancer cells, where we needed to preserve the natural state of the cells to assess differences accurately.
The wealth of data obtained from each run is quite substantial. Although the volume of data could be overwhelming, it allowed for extensive screening of multiple cellular readouts to comprehensively understand the drug’s impact on the cells. Moreover, the non-destructive nature of the HoloMonitor imaging facilitated the use of the same cells for follow-up assays. After visualization with the HoloMonitor, we were able to analyze these cells further using other techniques, such as flow cytometry and western blotting. This streamlined workflow was crucial for validating the imaging data and providing a more detailed molecular understanding of the cellular responses to the treatment.
Q3: In your recent publication1, you reported that Spinosyn A induced multiple morphological changes in T47-D breast cancer cells. What’s the importance of these morphological changes, and did these changes correlate with results on the molecular level?
A: In our recent publication on the effects of Spinosyn A (SPA) on T47-D breast cancer cells, we observed that SPA induced significant morphological changes, which are crucial for understanding its anticancer effects. These morphological alterations include changes in cell volume, shape, and motility, which are closely linked to the cells’ functional abilities and health. For example, increased cell volume and altered shape can indicate changes in the cell cycle and apoptosis, while modifications in cell motility can impact the cells’ ability to invade and metastasize.
These morphological changes observed through holographic imaging microscopy correlated with our molecular findings. The RNA-Seq analysis showed that SPA treatment altered the expression of 1380 genes involved in critical biological processes and signaling pathways such as cell proliferation, apoptosis, and the PI3K-Akt and MAPK pathways. For instance, changes in gene expression profiles that support reduced proliferation and increased apoptosis align with the observed increases in cell volume and roughness—a phenotype often associated with apoptotic cells.
Moreover, SPA’s impact on the MAPK and PI3K pathways, which are crucial for cell survival and growth, supports the observed morphological changes. These pathways’ modulation can lead to alterations in cell growth dynamics and structural integrity, further linking the observed morphological changes to SPA’s anticancer activities at a molecular level.
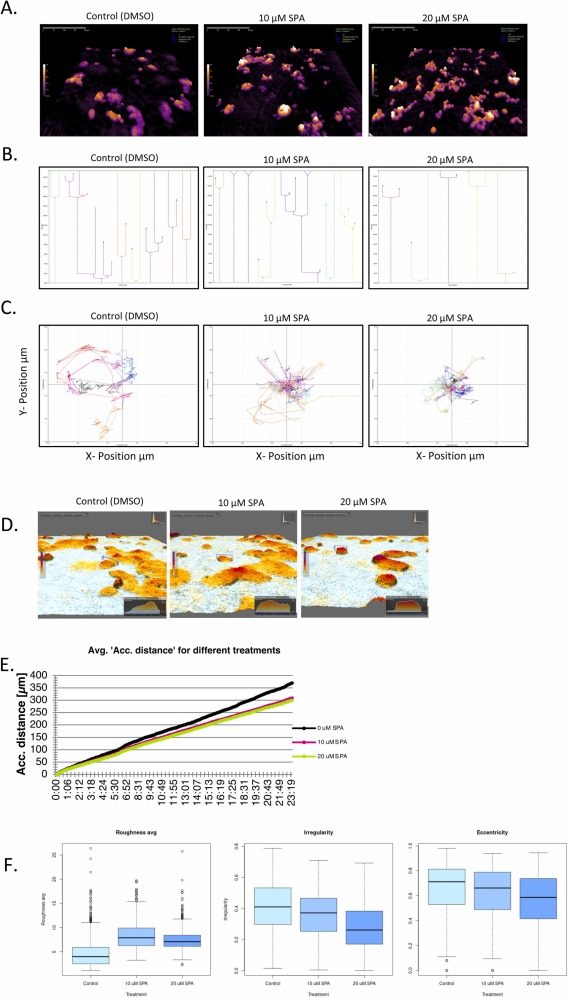
A. Live time lapse microscopy utilizing holographic monitoring imaging reveals that the cells are increasing in volume in a concentration-dependent manner when treated with SPA. The more yellow, the higher the cell height.
B, C. Single-cell tracking of cell families reveals that the cells are exhibiting longer division times, and their motility is reduced when exposed to SPA.
D, E. Peak measurements of the cell height and the accumulated distance of the cells measured utilizing holographic monitoring live imaging.
F. Various morphological features of the cells reveal that the cell roughness is increased in a concentration-dependent manner, whereas cell irregularity and eccentricity are decreasing. Kaniski et al. (2024)
Q4: In the same paper, you also used HoloMonitor to look at cell movement and cell division between control and treated cells on a single-cell level. Why do you think it is important to look at these data on a single cell level in addition to the population cell proliferation and motility data you already obtained?
A: Assessing cellular responses to treatments at the single-cell level, in addition to population-level data, provides critical insights that can enhance our understanding of drug effects on cellular heterogeneity. In cancer research, individual cells can respond differently to treatment due to inherent or acquired variability, which might be masked when looking only at population averages.
Using HoloMonitor’s single cell tracking ability, we can observe and quantify dynamic changes in individual cells in terms of movement and division over time. This allows us to capture a more detailed picture of how individual cells within a population respond to Spinosyn A.
For example, differences in cell cycle durations, migration patterns, and morphological changes can be crucial for understanding mechanisms of drug resistance or sensitivity at a single-cell level. Moreover, this approach helps identify subpopulations of cells that might behave differently, which is essential for developing more effective and personalized therapeutic strategies. Understanding these variations at the single-cell level enhances the overall interpretation of the cellular mechanisms at play, potentially leading to insights that could be overlooked by bulk analysis alone. This can enable better drug design and therapeutic approaches that target all cell types within a tumor, aiming for treatments that prevent escape mechanisms often exploited by a subset of cancer cells.
Q5: How has HoloMonitor benefited your research and workflow?
A: Integrating HoloMonitor into my workflow not only enhances the efficiency and depth of my experiments but also significantly conserves both time and financial resources. Given that I am operating within a smaller institution, it’s crucial to optimize each aspect of our research. HoloMonitor’s capability to provide valuable, real-time data without the need for additional reagents or preparations—such as dyes or labels—means that we can conduct non-invasive, live-cell imaging more cost-effectively.
This efficiency is particularly advantageous because it allows for the continuous monitoring of cell health and behavior without the additional costs associated with consumables typically required for traditional cell imaging techniques. By reducing the need for these consumables, we save on recurring expenses and minimize our laboratory waste, aligning with both budgetary and environmental considerations.
Moreover, the time saved by using HoloMonitor is substantial. Traditional methods often require extensive preparation and processing time, from staining cells to acquiring and processing images. With HoloMonitor, we can set up experiments quickly and collect data continuously without manual intervention, freeing up time to focus on data analysis and other research activities. This streamlining of processes significantly accelerates our research, allowing us to achieve results faster and more efficiently. This aspect of HoloMonitor is especially valuable in a high-throughput setting, where we can screen multiple conditions or treatments concurrently, further maximizing our research capabilities within the constraints of a smaller institution.
Q6: How do you best utilize the vast amount of information that the HoloMonitor provides? Can you tell us more about the AI tools you are using to help you in your data analytics and preparation?
To effectively utilize the vast amount of data provided by the M4 HoloMonitor, I integrate AI tools developed by Dr. Xijin “Steven” Ge, focusing on RTutor and Chatlize.ai. These tools support advanced data analysis, crucial for managing the complex datasets generated by HoloMonitor.
Chat with your data
Dr. Xijin “Steven” Ge, Professor of Bioinformatics at South Dakota State University, holds a PhD from The University of Tokyo and is known for his work on artificial neural networks. His NIH-supported research focuses on developing bioinformatics tools like RTutor and Chatlize.ai, designed to make complex data analysis more accessible, contributing significantly to genomics and bioinformatics.
RTutor is an innovative AI-powered application that translates plain English into executable R or Python code, streamlining data processing and analysis from HoloMonitor to discern cellular dynamics and responses quickly. Chatlize.ai, another of Dr. Ge’s innovations, enhances data interaction through conversational AI, enabling intuitive data exploration that is essential in fast-paced research environments where quick decision-making is paramount.
By incorporating these AI tools into our workflow, we enhance our understanding of cellular behaviors and treatment effects, using cutting-edge technology to improve research outcomes. This approach is especially valuable for institutions with limited budgets, as it ensures that advanced analytical capabilities remain within reach, fostering continued innovation in research.
Dr. Ge introduces Chatlize.ai
Dr. Ge explains how to talk to your data with RTutor.ai
Q7: If other users were interested in following your example of leveraging AI to analyze their HoloMonitor data, what coding experience is required, and what advice would you have?
For users interested in leveraging AI tools like RTutor and Chatlize.ai to analyze HoloMonitor data, the great news is that extensive coding experience is not required. These tools are designed to be user-friendly and accessible, allowing you to interact with and manipulate data using plain English. This means you can command the software through simple human language to perform complex data analysis tasks without needing to write code.
For those who are more comfortable with programming, these tools also offer the flexibility to take the generated code and further customize it in Python or R. This adaptability caters to users with varying levels of programming proficiency, from beginners to experienced coders.
My advice to new users would be to start by familiarizing themselves with the basic functions of RTutor and Chatlize.ai, utilizing the intuitive interfaces these tools provide. Since I started using these applications, I haven’t needed to manually code for my analyses because the outputs, including the graphics, are comprehensive, visually appealing, and ready for publication.
In summary, whether you’re new to coding or an experienced programmer, these tools can significantly streamline your data analysis process, making your research more efficient and effective.
Q8: Looking ahead, how do you anticipate using HoloMonitor in your future research projects?
Additionally, I aim to integrate HoloMonitor data with multi-omics analyses to deepen our insights into how cells respond to therapies at the molecular level. This approach will enhance our understanding of cancer biology and aid in developing more effective therapeutic strategies.
Q9: Which aspects of the system do you find most valuable for your work?
One of the system’s most valuable aspects for my work is its cost-effectiveness. This affordability allows for continuous and extensive data collection and analysis, making it an indispensable tool in my research for studying cellular behaviors and treatment responses.
I can use HoloMonitor all day, every day, without additional costs—well, except for maybe splurging on extra hard drive space!
Q10: Considering the wide range of HoloMonitor’s capabilities, which fields of study do you believe would benefit most from integrating this technology into their experiments and why?
I think HoloMonitor’s versatile capabilities make it highly valuable across various scientific fields. Integrating the HoloMonitor into their experiments enables researchers to achieve more precise, real-time observations that enhance the depth and quality of scientific inquiry.
In cancer research, for instance, its ability to monitor real-time cell motility, proliferation, and morphological (and many other) changes is crucial for understanding tumor progression and treatment responses.
The system is also highly beneficial in drug discovery and development, enabling researchers to assess the efficacy and toxicity of new drug candidates efficiently.
In the realm of stem cell research, the non-invasive imaging capabilities of the HoloMonitor allow for the observation of differentiation and growth, which are critical in regenerative medicine.
Additionally, in immunology, the HoloMonitor can track immune cell interactions, providing detailed insights into immune responses and aiding in the development of new immunotherapies.
Developmental biologists can also leverage this technology to study cell development and differentiation processes, offering a granular view of cellular changes over time.
Reference:
- Kaniski et al. (2024) Spinosyn A exerts anti-tumorigenic effects on progesterone-sensitive ERα-positive breast cancer cells by modulating multiple signaling pathways, Biomedicine & Pharmacotherapy 171 (2024) 116156, https://doi.org/10.1038/s41598-020-71538-1.
Thank you to Besa for this interview!