Single Cell Tracking Assay Protocol

This protocol helps to set up a Single Cell Tracking using HoloMonitor® M4 and the HoloMonitor® App Suite software. The HoloMonitor® Single Cell Tracking facilitates label-free characterization of heterogeneous live cell behavior overtime on an individual cell level. The HoloMonitor® Single Cell Tracking can also be used to analyse fluorescence data obtained with HoloMonitor® M4FL, for more information see HoloMonitor® Fluorescence Capture protocol.
Requirements:
- HoloMonitor® M4, placed in incubator
- HoloMonitor® M4 App Suite
- Culture vessel of choice with cells
- HoloLid™ for selected vessel
- Vessel holder for selected vessel
Output:
- Plots of cell movement and morphological features over time
- Interactive cell family trees
- Calculation spread sheets of all data including graphs and population averages
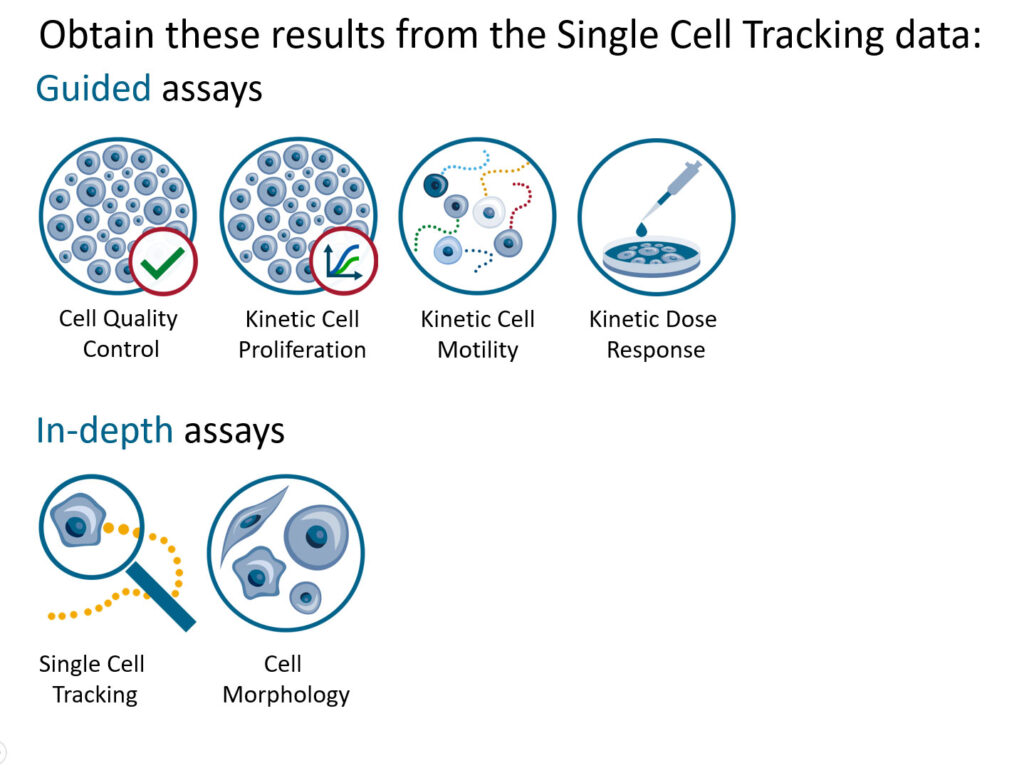
Reanalysis:
- Guided assays
- In-depth analysis
PREPARATIONS
Materials:
- HoloMonitor® M4, placed inside the incubator
- HoloMonitor® App Suite software
- Cell culture vessel. Please check our list of recommended vessels.
- HoloLid™ for the selected vessel
- Vessel holder for selected vessel
- Setup and Operation Manual for HoloMonitor® M4
Steps
Seed the cells with about 5 % confluence (ca. 6000 – 11000 cells/cm2).
►Please note that too few cells may lead to inadequate results due to auto-focus failure.
Place the vessel in the incubator and let cells attach for 2-24 hours.
Start the software and wait for complete instrument initialization.
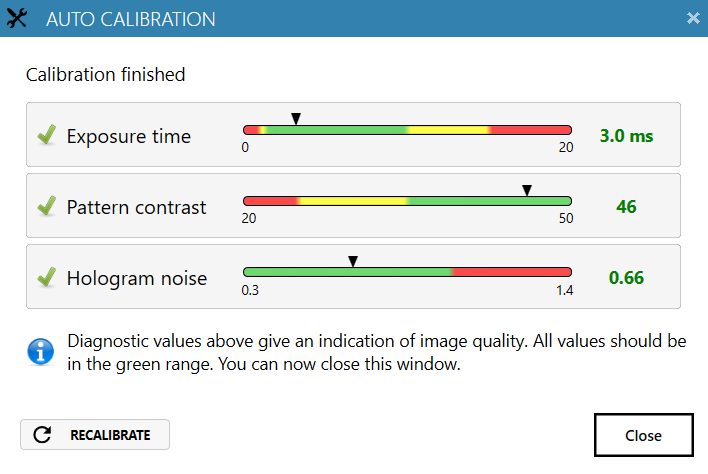
Run an auto-calibration. With successful calibration (as shown in the picture), the instrument is ready to use.
| Vessel |
Vendor cat. number | HoloLid™ |
Final working volume |
Growth area, cm2/well |
Vessel cut in a holder |
| Sarstedt TC-dish 35 | 83.3900 | 71110 | 3.0 mL/well | 8.00 | NA |
| Sarstedt TC 6-well plate | 83.3920.005 | 71120 | 3.0 mL/well | 8.80 | top left |
| Sarstedt lumox® 24-multiwell plate | 94.6000.014 | 71130 | 1.9 mL/well | 1.90 | top left |
| Sarstedt lumox® 96-multiwell plate | 94.6000.024 | 71140 | 170 µL/well | 0.34 | top left |
| ibidi® μ-dish 35 mm, high | 81156 | 71111 | 2.5 mL/well | 3.50 | NA |
| ibidi® μ-plate 24 Well Black | 80241 | 71131 | 2.5 mL/well | 1.90 | NA |
| Eppendorf CCCadvanced® FN1 – 6 well | 0038110010 | 71150 | 3.0 mL/well | 9.40 | bottom right |
Sterilize the HoloLids™ according to the HoloLid™ sterilization and use protocol.
Add the treatment to your cells. The final working volumes per well, essential for using HoloLids™, are shown in the table above.
Slide the cell culture vessel onto the Vessel holder, its grips facing towards you. Ensure that the vessel is parallel to the holder. There is a spring that holds the vessel in place.
►When using multi-well plates, place them with the cut-off corner to the left.
Replace the standard lids with the HoloLid™.
Put the vessel holder with the sample on the HoloMonitor® M4 stage.
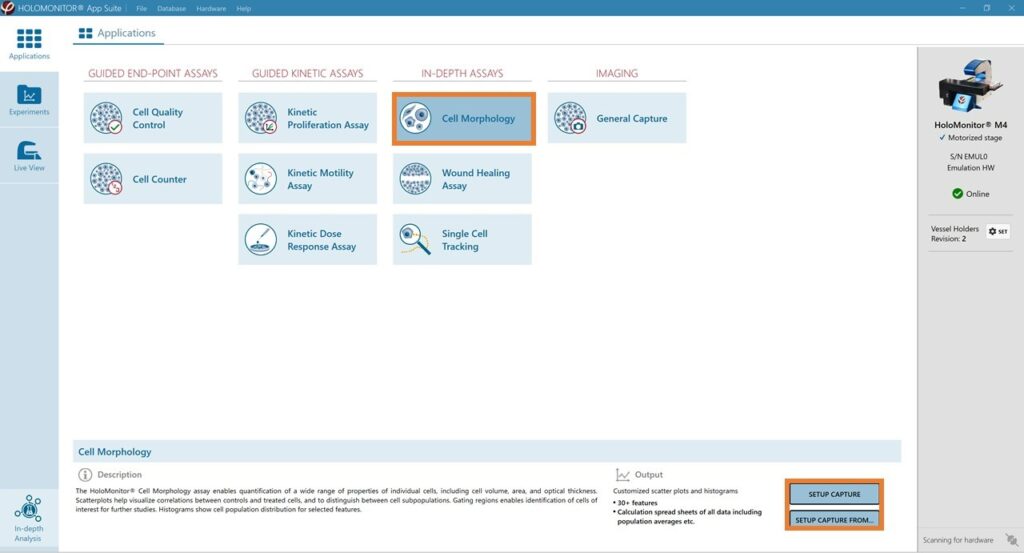
Select the Single Cell Tracking Assay and proceed by clicking the Setup Capture button.
EXPERIMENT SETUP
Basic setup: Describe the experiment and assign treatments to the wells
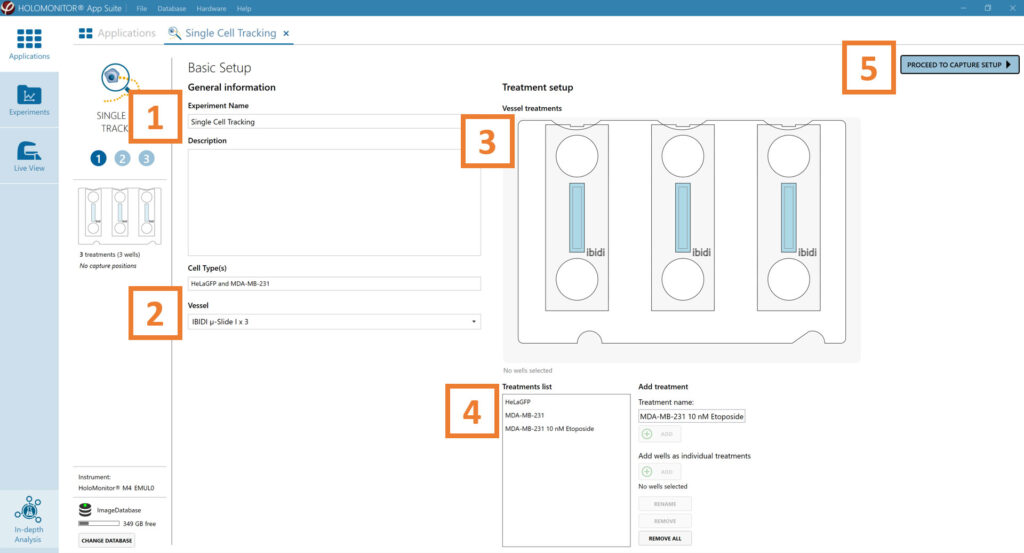
Enter the experiment name, optional experiment description, and cell types.
Select the correct vessel map from the drop-down list.
Map treatments and conditions on the vessel map. Select wells by marking them with the left mouse button while moving the cursor over the relevant well/s.
Add the treatment name(s) in the text box below the vessel map and click Add /press Enter. It is possible to add wells as individual treatments. Marked well/s are light blue, selected wells will appear dark blue.
Proceed to Capture setup.
Capture setup: Select the experiment time settings and choose capture positions
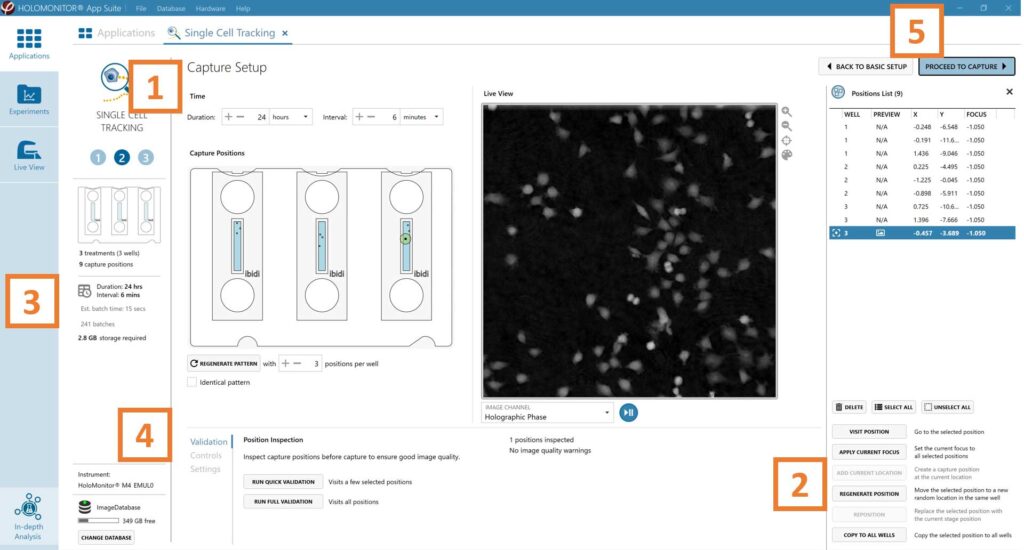
Adjust the default settings for duration and interval.
Add capture positions: The position list is open by default. Click positions on the vessel map and add them to the position list with the Add current location button. In case the image quality is poor, a warning sign appears. Adjust focus or position location if necessary.
Ensure that the storage requirement for the experiment does not exceed the computer capacity.
► When running an experiment, data needs to be stored on the computer connected to the instrument. Storing data on an external drive (e.g. connected via USB or internet server) may cause data loss due to erratic USB connections or poor internet connection.
Run a full or quick validation of the selected positions to ensure good image quality.
When satisfied with the experiment setup, click Proceed to Capture.
Capture: Review the experiment in real-time during the time-lapse
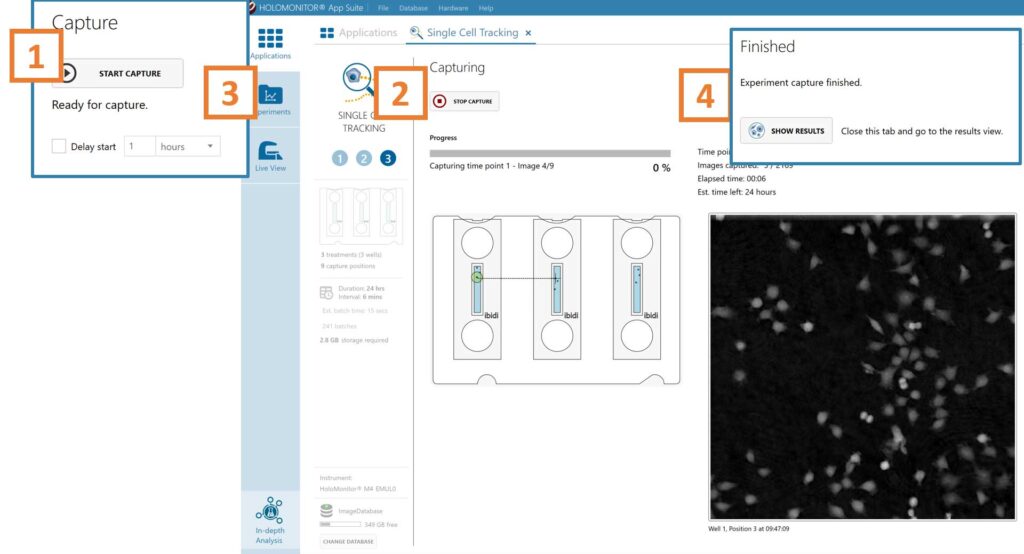
Click Start Capture.
To stop the experiment ahead of time, click the stop button.
► Note that it is NOT possible to restart the experiment once it has been stopped.
Go to the Experiments tab and open your ongoing experiment to preview the results during the run.
► Wait for the experiment to finish before starting the In-depth Analysis.
When the Experiment capture finishes, click the Show Result button to get directly to the Results page.
RESULTS & ANALYSIS
Experiments tab
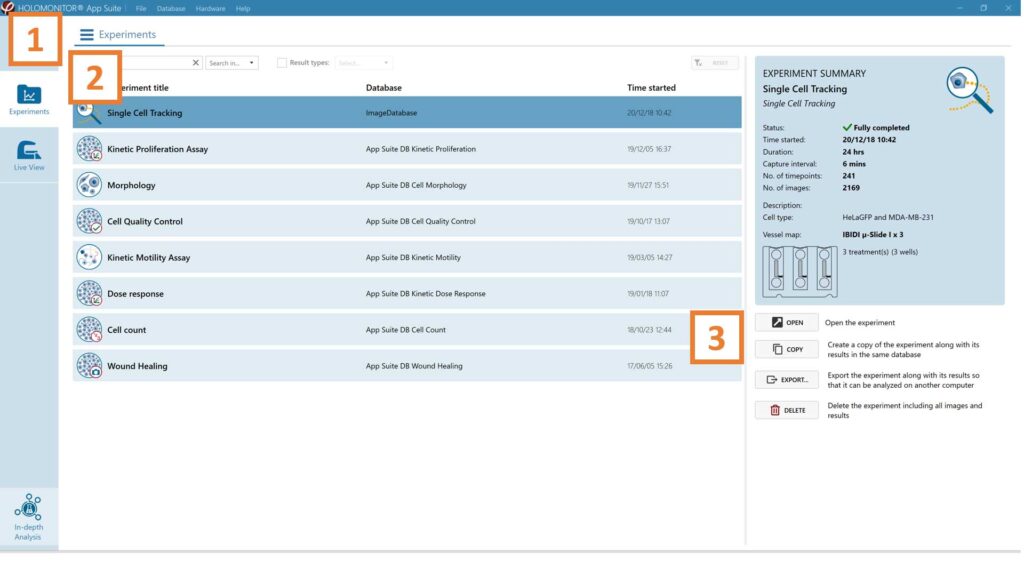
Click Experiments to see a list of the experiments.
Click on the experiment title to open an experiment summary.
Click Open to open the results page.
Experiment overview tab
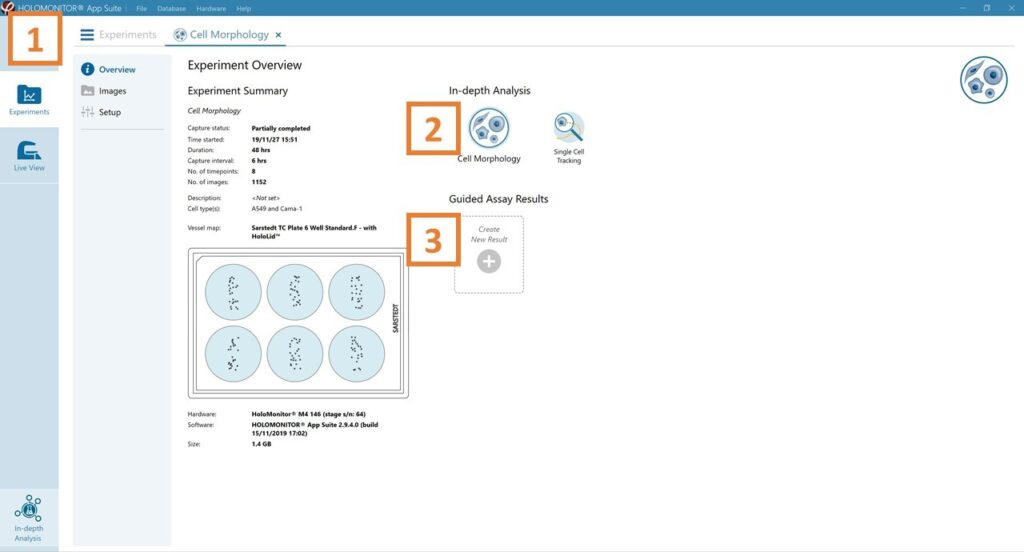
See the experiment summary, view all images, and go to the experiment setup by choosing the respective tab.
Generate in-depth analysis data from the captured images by clicking on the Single Cell Tracking icon. A new window for in-depth analysis will open.
Create New Guided Assay Results from this experiment by clicking the respective button.
In-depth analysis — identify cells tab
- First step for morphology analysis is to identify the cells.
- The software automatically calculates cell count and confluence of each image.
- Review the image quality and include/ exclude images from results analysis. See the Image quality guide for more information.
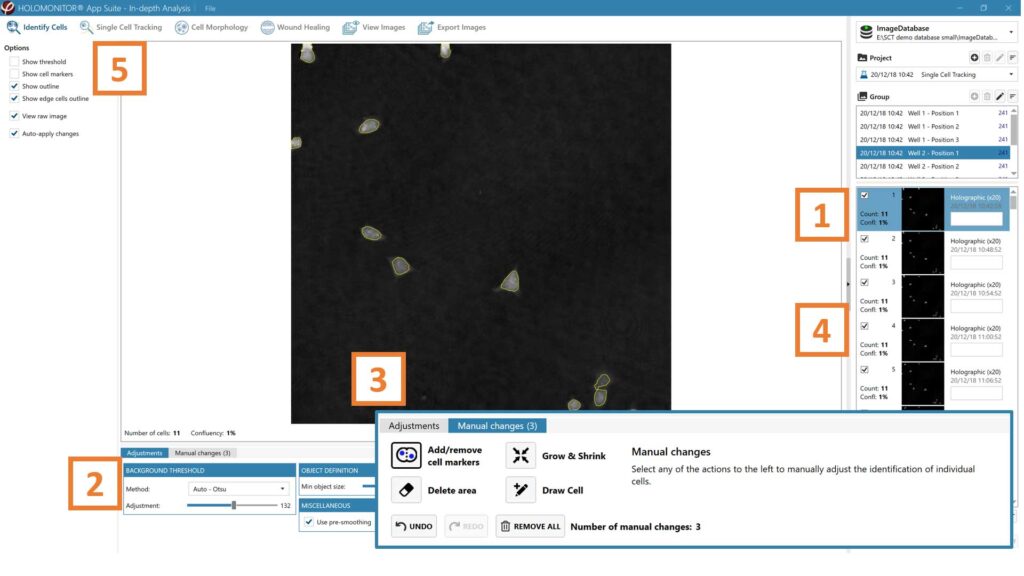
It is the users’ responsibility to verify that the software algorithm was correctly applied during analysis and that results are accurate.
Start by selecting a frame of interest, so the image is displayed. You can alter the viewing options.
Adjust the background threshold and object size so the mask best fits the cell segmentation and click. Apply (Current/ Checked/ All) to fix the settings to the frames and to display the automatic calculation of cell count (#/frame) and confluence (%).
Manual changes are possible. Add/remove or grow/ shrink a cell, delete a cell area or draw a cell area yourself. These changes will only be saved to the currently selected image.
Check cell segmentation for some images in the beginning, middle, and end of the experiment to make sure the applied threshold fits. Scrolling through the image list and checking for abnormal jumps in cell count or confluence values may help as well to see if adjustments are necessary.
Once satisfied with the cell identification, click on the Single Cell Tracking tab to begin analysis.
In-depth analysis — Wound healing tab
- Review the image quality and include/ exclude images from results analysis. See the Image quality guide for more information.
- For further analysis all results can be exported.
- For more details, see the Setup and Operation manual.
Click New Analysis to start a new tracking analysis or
load a previous one by clicking the Open button.
• Always start resolving warnings by adjusting cell identification. If this step fails, only then adjust tracking manually.
• Use the Auto Track function after manual track adjustments.
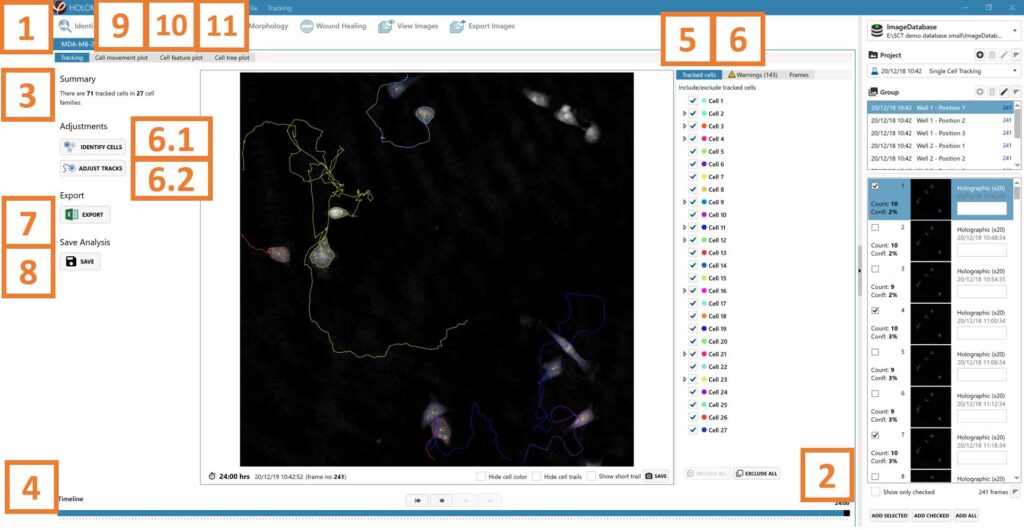
Add at least two frames to the analysis using the Add Selected/ Add Checked/ Add All button.
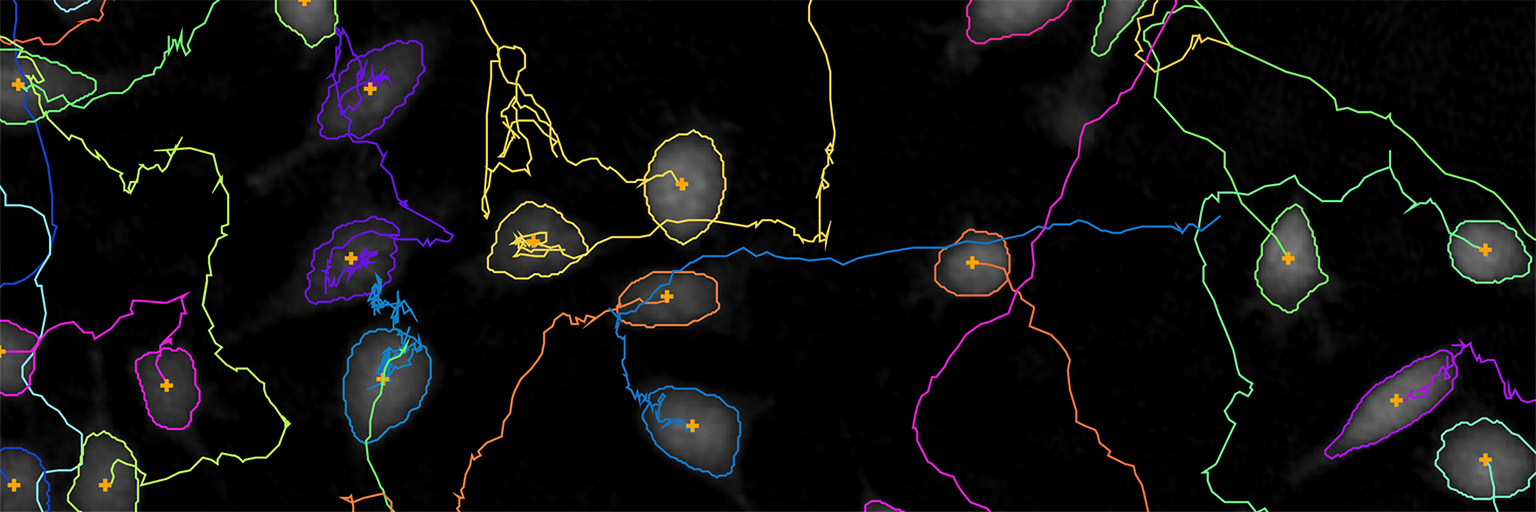
The software automatically tracks all the cells in the included frames from a time-lapse.
► At the top left corner, there is a summary of the number of cell and cell families tracked.
Use the timeline slider to view cell tracks over time in the time-lapse and evaluate cell tracking status.
Include and exclude cells and/or cell families in the tracking by checking/unchecking tick boxes in the tracked cells list.
► Tracking information and cell features for cells and/or cell families included in tracked cells list will be added to the Excel export file.
A list of possible warnings resulting from tracking issues such as cells leaving the field of view, overlapping, or dividing. Resolve the warnings by:
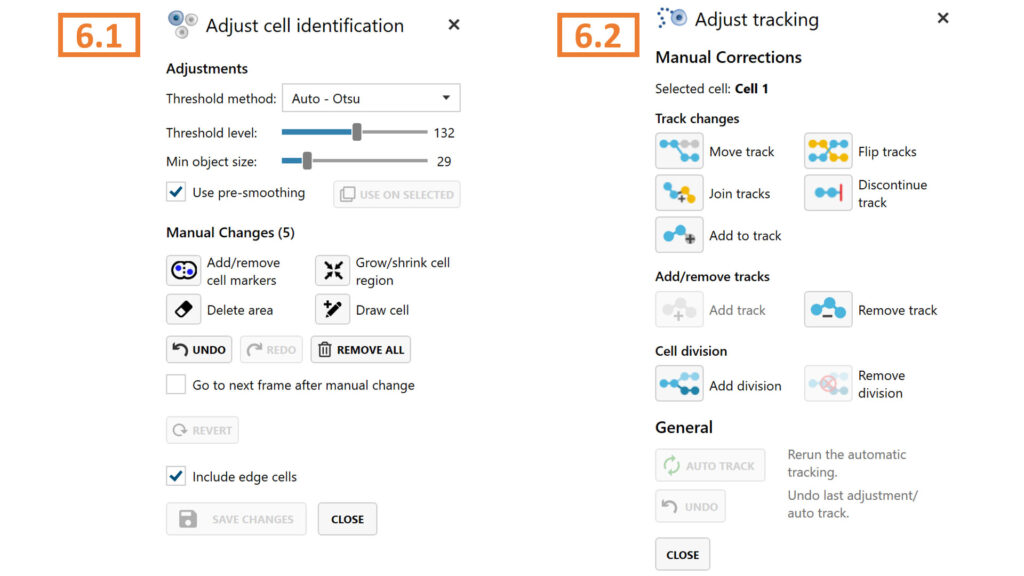
6.1. first adjusting specific cell and or full-frame segmentation using Identify cells functions.
► Segmentation changes created here are applied globally and can influence the results in other In-depth assays.
6.2. then adjusting tracks manually. For more details on each button function, see the Setup and Operation manual, or choose to ignore the warning.
► Repeat steps 6.1. to 6.3. until all warnings are resolved and cell tracks are correct.
Export the tracking data to Excel. Cell features for cells and cell families included in step 5 are exported. The exported file can include cell feature averages, cell feature data for all individually tracked cells and cell families, as well as cell movement and cell tree plots.
Save tracking analysis to return to it later.
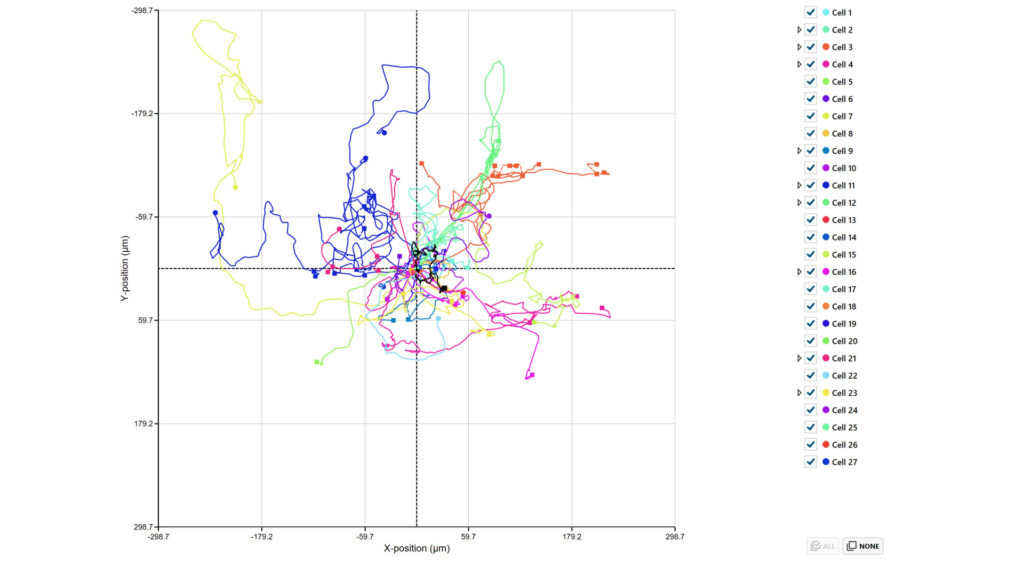
In the Cell movement plot tab, the tracking data for the selected cells and/or cell families are displayed in a plot showing the cell track traces. Plots can be saved and scale/display options can be changed.
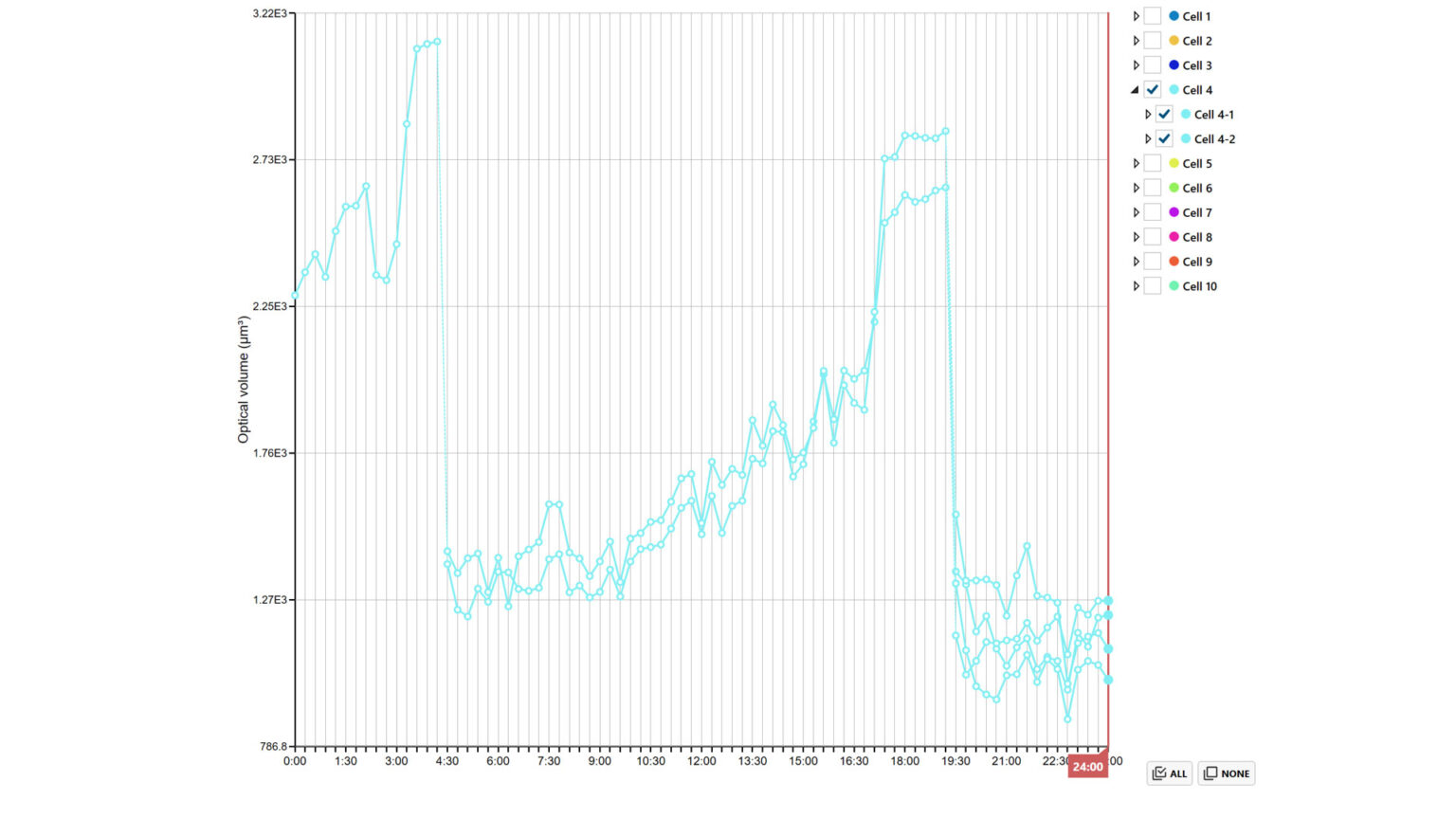
In the Cell feature plot tab, cell feature changes of selected cells and/or cell families can be viewed over time. Select the feature of interest from the drop-down list. Plots can be saved and scale/display options can be changed.
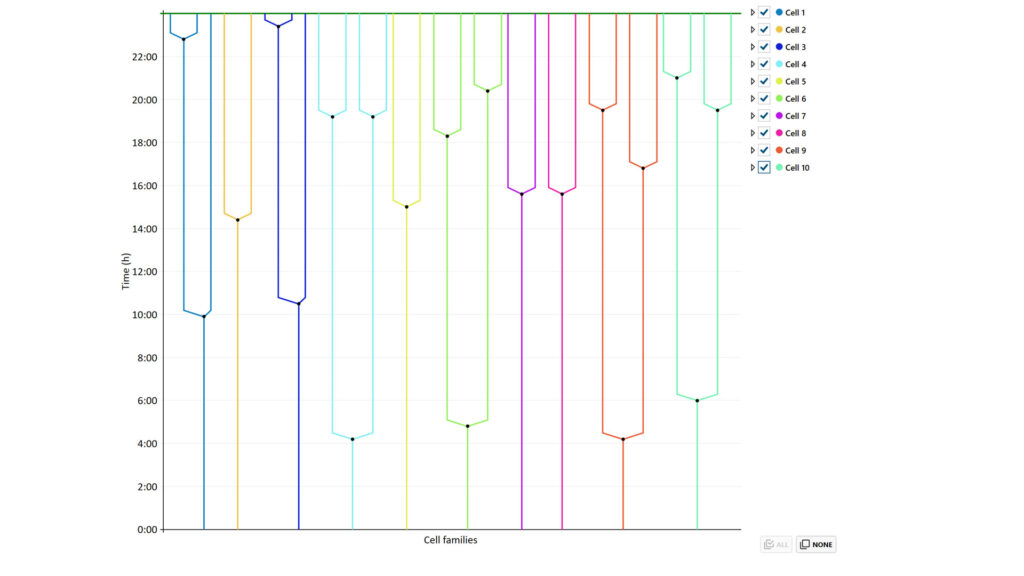
In the Cell tree plot tab, cell family trees for selected cell families feature changes of selected cell families are shown. Plots can be saved and scale/display options can be changed.
One experiment — multiple results
- This section helps to reanalyze data between different assays using HoloMonitor® App Suite software.
Generating In-depth Assay results
Generating Guided Assay results
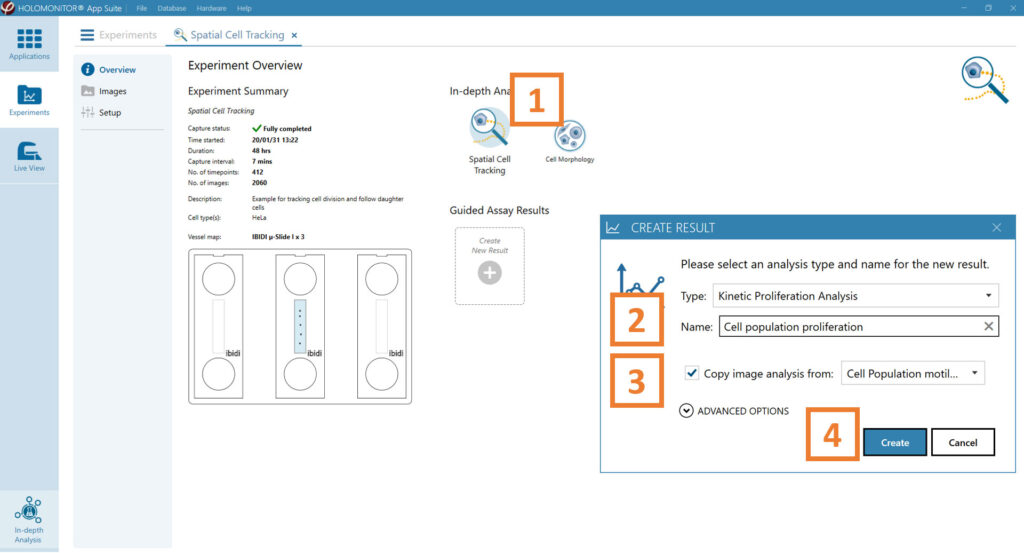
In the Experiment overview page under Guided Assay Results select Create New Result.
Choose the type of analysis in the pop-up window and name the new result.
Tick copy image analysis from and select the experiment to copy from. This will copy the image analysis settings from the selected result including all changes.
► For further data analysis steps, please see the respective assay protocol.
Press create.